
rxode2 Transit Compartment Models
2026-02-17
Source:vignettes/rxode2-transit-compartments.Rmd
rxode2-transit-compartments.RmdSavic 2008 first introduced the idea of transit compartments being a mechanistic explanation of a a lag-time type phenomena. rxode2 has special handling of these models:
You can specify this in a similar manner as the original paper. Note
that we use evid=7 instead of evid=1 for these
sorts of transit compartment models. evid=7 is the transit
compartment model/phantom event. This puts the dose in the
dose() function and calculates time since last dose
tad() but doesn’t actually put the dose in the compartment.
This allows the transit() function to easily apply to the
compartment.
## rxode2 5.0.1.9000 using 2 threads (see ?getRxThreads)
## no cache: create with `rxCreateCache()`
mod <- function() {
model({
## Table 3 from Savic 2007
cl = 17.2 # (L/hr)
vc = 45.1 # L
ka = 0.38 # 1/hr
mtt = 0.37 # hr
bio=1
n = 20.1
k = cl/vc
ktr = (n+1)/mtt
## note that lgammafn is the same as lgamma in R.
d/dt(depot) = exp(log(bio*podo(depot))+log(ktr)+n*log(ktr*tad(depot))-
ktr*tad(depot)-lgammafn(n+1))-ka*depot
d/dt(cen) = ka*depot-k*cen
})
}
et <- et(0, 7, length.out=200) |>
et(amt=20, time=0, evid=7)
transit <- rxSolve(mod, et)## i parameter labels from comments are typically ignored in non-interactive mode## i Need to run with the source intact to parse comments
plot(transit, cen, ylab="Central Concentration")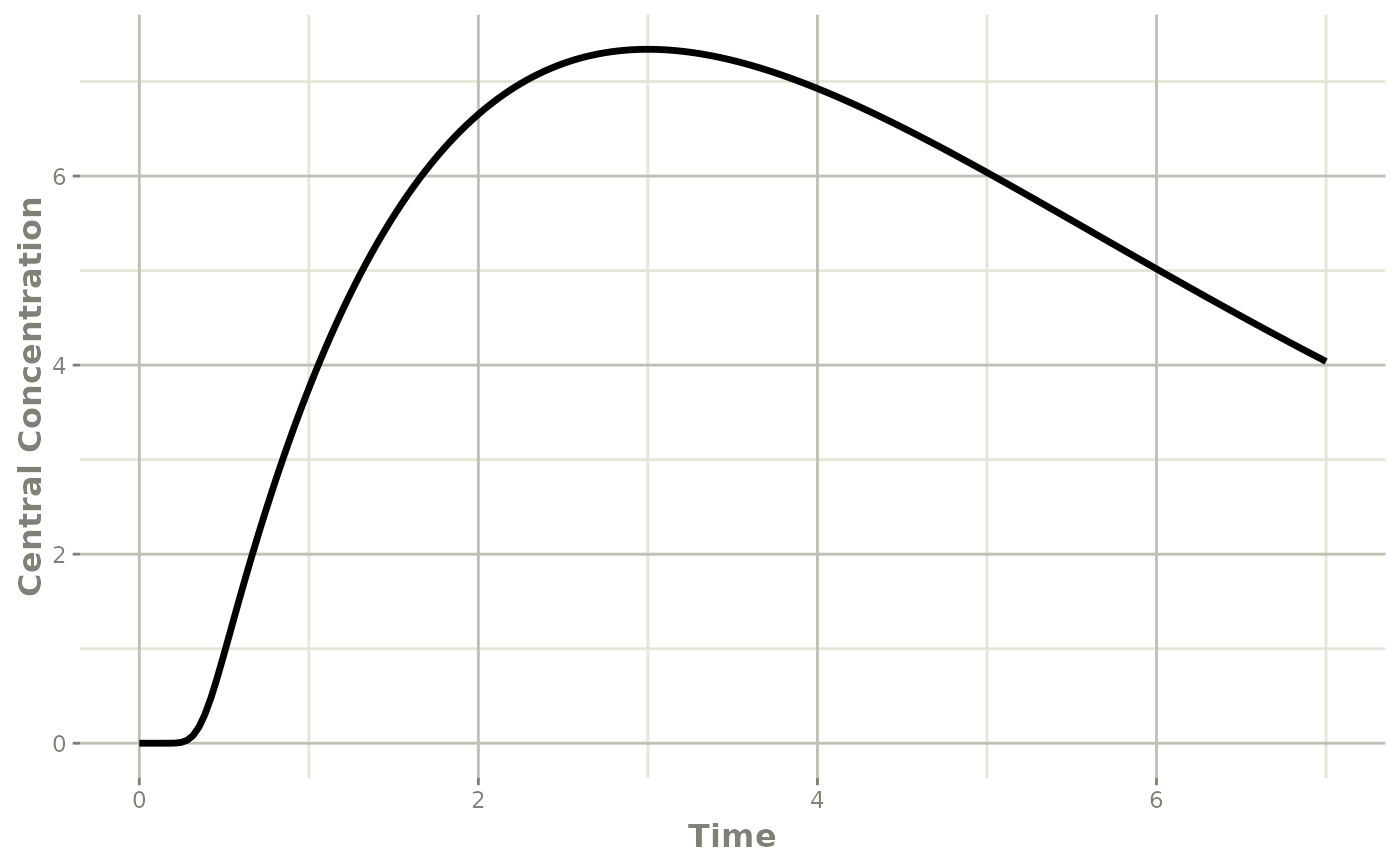
Another option is to specify the transit compartment function
transit syntax. This specifies the parameters
transit(number of transit compartments, mean transit time, bioavailability).
The bioavailability term is optional.
The same model can be specified by:
mod <- function() {
ini({
## Table 3 from Savic 2007
cl <- 17.2 # (L/hr)
vc <- 45.1 # L
ka <- 0.38 # 1/hr
mtt <- 0.37 # hr
bio <- 1
n <- 20.1
})
model({
k <- cl/vc
ktr <- (n+1)/mtt
d/dt(depot) <- transit(n,mtt,bio)-ka*depot
d/dt(cen) <- ka*depot-k*cen
})
}
et <- et(0, 7, length.out=200) |>
et(amt=20, evid=7)
transit <- rxSolve(mod, et)## i parameter labels from comments are typically ignored in non-interactive mode## i Need to run with the source intact to parse comments
plot(transit, cen, ylab="Central Concentration")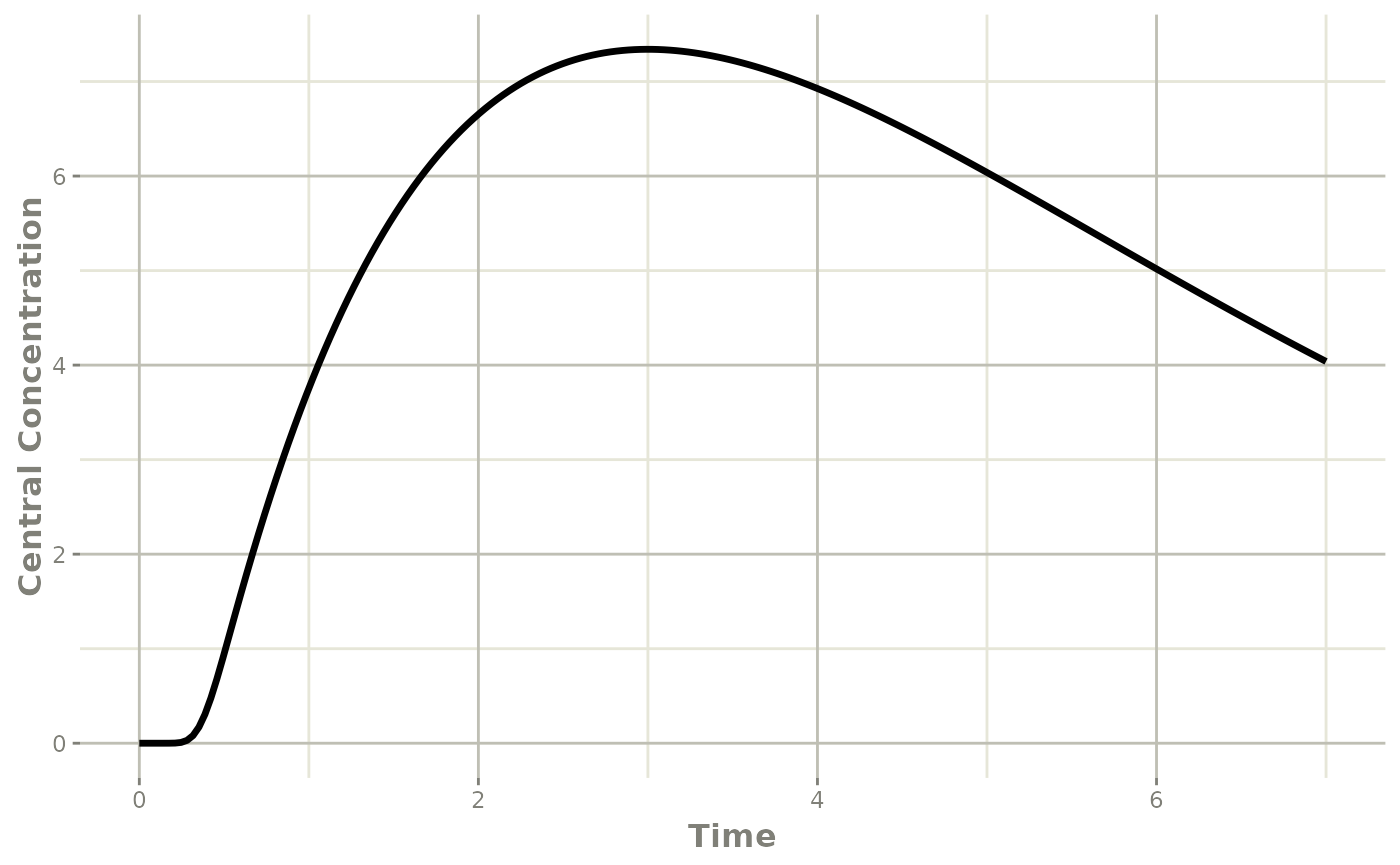
A couple of things to keep in mind when using this approach:
This approach implicitly assumes that the absorption through the transit compartment is completed before the next dose begins
Different types of doses (ie bolus/infusion) to the compartment affect the time after dose calculation (
tad) which is used in the transit compartment calculation. These (therefore) are not currently supported. The most stable way is to usetad(cmt)andpodo(cmt), this way doses to other compartments do not affect the transit compartment calculation.Internally, the
transitsyntax uses either the currently defined cmtd/dt(cmt)=transit(...), orcmt. If the transit compartment is used outside of ad/dt()(not recommended), thecmtthat is used is the lastd/dt(cmt)defined it the model. This also means compartments do not affect one another (ie a oral, transit compartment drug dosed immediately with an IV infusion)