Introduction
rxode2 is an R package that facilitates simulation with
ODE models in R. It is designed with pharmacometrics models in mind, but
can be applied more generally to any ODE model.
Description of rxode2 illustrated through an example
The model equations can be specified through a text string, a model
file or an R expression. Both differential and algebraic equations are
permitted. Differential equations are specified by
d/dt(var_name) =. Each equation can be separated by a
semicolon.
To load rxode2 package and compile the model:
library(rxode2)
#> rxode2 5.0.1.9000 using 2 threads (see ?getRxThreads)
#> no cache: create with `rxCreateCache()`
mod1 <- function() {
ini({
# central
KA=2.94E-01
CL=1.86E+01
V2=4.02E+01
# peripheral
Q=1.05E+01
V3=2.97E+02
# effects
Kin=1
Kout=1
EC50=200
})
model({
C2 <- centr/V2
C3 <- peri/V3
d/dt(depot) <- -KA*depot
d/dt(centr) <- KA*depot - CL*C2 - Q*C2 + Q*C3
d/dt(peri) <- Q*C2 - Q*C3
eff(0) <- 1
d/dt(eff) <- Kin - Kout*(1-C2/(EC50+C2))*eff
})
}Model parameters may be specified in the ini({}) model
block, initial conditions can be specified within the model with the
cmt(0)= X, like in this model
eff(0) <- 1.
You may also specify between subject variability initial conditions
and residual error components just like nlmixr2. This allows a single
interface for nlmixr2/rxode2 models. Also
note, the classic rxode2 interface still works just like it
did in the past (so don’t worry about breaking code at this time).
In fact, you can get the classic rxode2 model
$simulationModel in the ui object:
mod1 <- mod1() # create the ui object (can also use `rxode2(mod1)`)
mod1
summary(mod1$simulationModel)Specify Dosing and sampling in rxode2
rxode2 provides a simple and very flexible way to
specify dosing and sampling through functions that generate an event
table. First, an empty event table is generated through the “et()”
function. This has an interface that is similar to NONMEM event
tables:
ev <- et(amountUnits="mg", timeUnits="hours") |>
et(amt=10000, addl=9,ii=12,cmt="depot") |>
et(time=120, amt=2000, addl=4, ii=14, cmt="depot") |>
et(0:240) # Add samplingYou can see from the above code, you can dose to the compartment named in the rxode2 model. This slight deviation from NONMEM can reduce the need for compartment renumbering.
These events can also be combined and expanded (to multi-subject
events and complex regimens) with rbind, c,
seq, and rep. For more information about
creating complex dosing regimens using rxode2 see the rxode2
events vignette.
Solving ODEs
The ODE can now be solved using rxSolve:
x <- mod1 |> rxSolve(ev)
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
x
#> ── Solved rxode2 object ──
#> ── Parameters (x$params): ──
#> KA CL V2 Q V3 Kin Kout EC50
#> 0.294 18.600 40.200 10.500 297.000 1.000 1.000 200.000
#> ── Initial Conditions (x$inits): ──
#> depot centr peri eff
#> 0 0 0 1
#> ── First part of data (object): ──
#> # A tibble: 241 × 7
#> time C2 C3 depot centr peri eff
#> [h] <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0 0 0 10000 0 0 1
#> 2 1 44.4 0.920 7453. 1784. 273. 1.08
#> 3 2 54.9 2.67 5554. 2206. 794. 1.18
#> 4 3 51.9 4.46 4140. 2087. 1324. 1.23
#> 5 4 44.5 5.98 3085. 1789. 1776. 1.23
#> 6 5 36.5 7.18 2299. 1467. 2132. 1.21
#> # ℹ 235 more rowsThis returns a modified data frame. You can see the compartment values in the plot below:
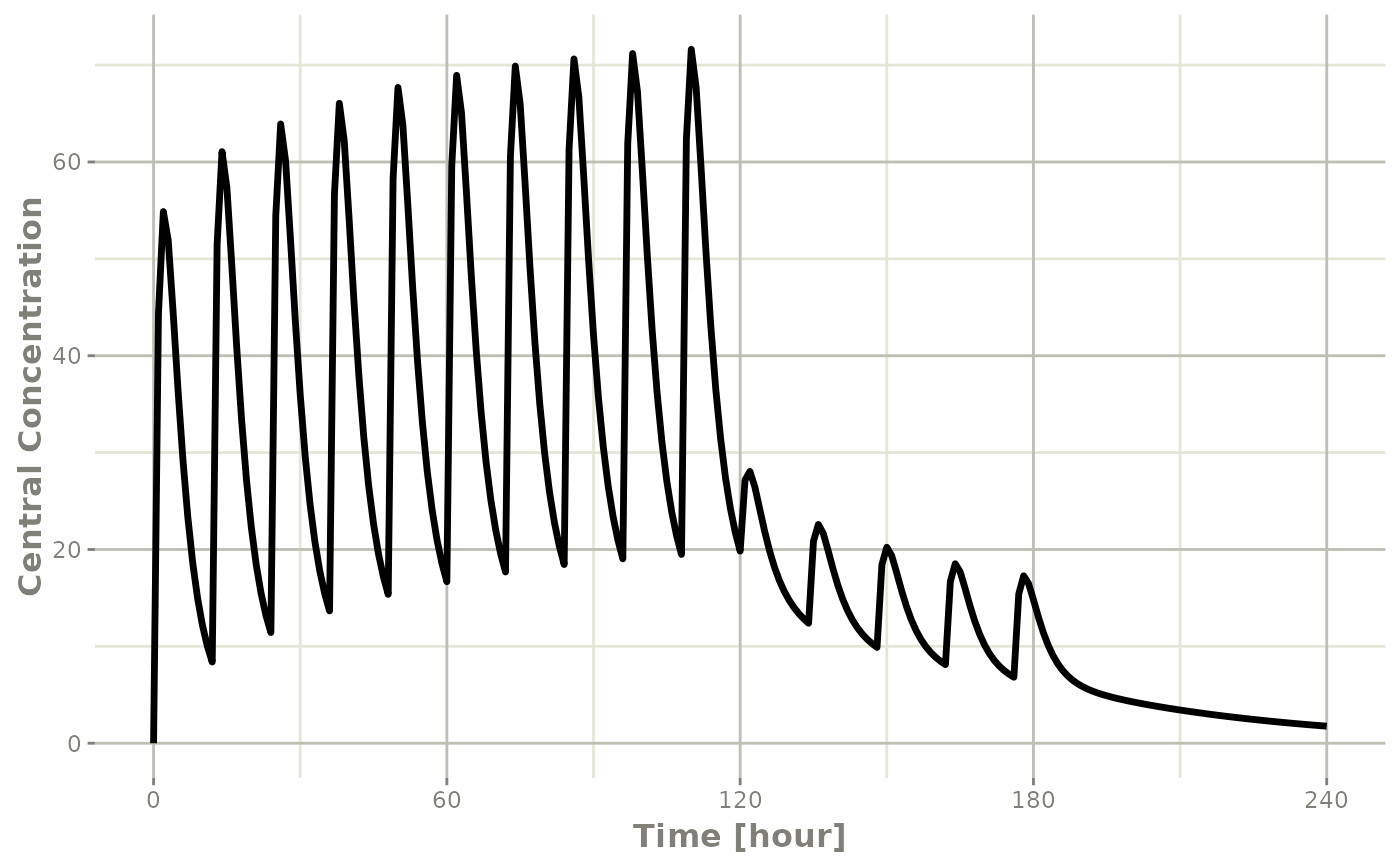
Or,
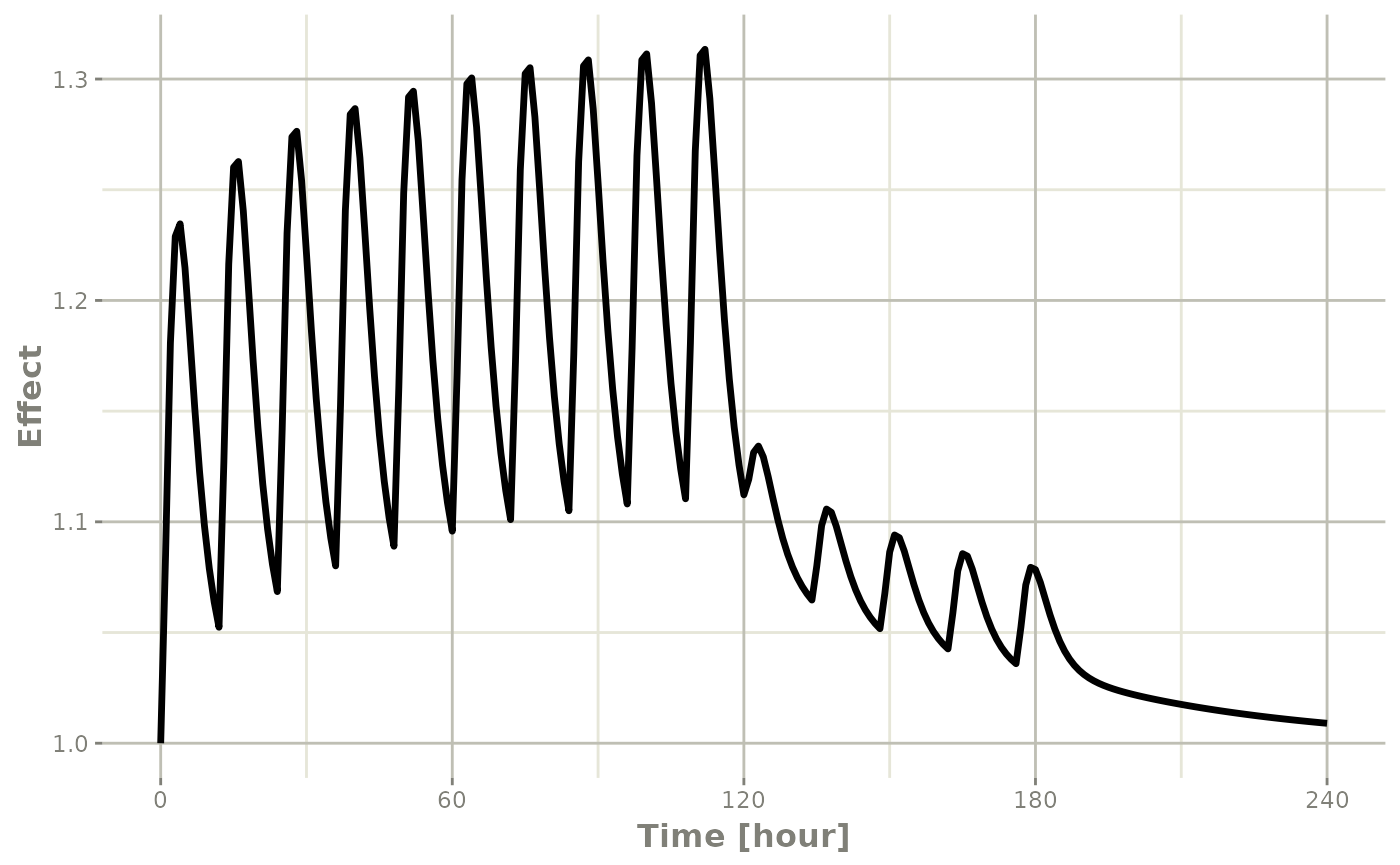
Note that the labels are automatically labeled with the units from
the initial event table. rxode2 extracts units to label the
plot (if they are present).
