VPC based on ui model
Usage
vpcPlot(
fit,
data = NULL,
n = 300,
bins = "jenks",
n_bins = "auto",
bin_mid = "mean",
show = NULL,
stratify = NULL,
pred_corr = FALSE,
pred_corr_lower_bnd = 0,
pi = c(0.05, 0.95),
ci = c(0.05, 0.95),
uloq = fit$dataUloq,
lloq = fit$dataLloq,
log_y = FALSE,
log_y_min = 0.001,
xlab = NULL,
ylab = NULL,
title = NULL,
smooth = TRUE,
vpc_theme = NULL,
facet = "wrap",
scales = "fixed",
labeller = NULL,
vpcdb = FALSE,
verbose = FALSE,
...,
seed = 1009,
idv = "time",
cens = FALSE,
method = c("vpc", "tidyvpc")
)
vpcPlotTad(..., idv = "tad")
vpcCensTad(..., cens = TRUE, idv = "tad")
vpcCens(..., cens = TRUE, idv = "time")Arguments
- fit
nlmixr2 fit object
- data
this is the data to use to augment the VPC fit. By default is the fitted data, (can be retrieved by
getData), but it can be changed by specifying this argument.- n
Number of VPC simulations
- bins
either "density", "time", or "data", "none", or one of the approaches available in classInterval() such as "jenks" (default) or "pretty", or a numeric vector specifying the bin separators.
- n_bins
when using the "auto" binning method, what number of bins to aim for
- bin_mid
either "mean" for the mean of all timepoints (default) or "middle" to use the average of the bin boundaries.
- show
what to show in VPC (obs_dv, obs_ci, pi, pi_as_area, pi_ci, obs_median, sim_median, sim_median_ci)
- stratify
character vector of stratification variables. Only 1 or 2 stratification variables can be supplied.
- pred_corr
perform prediction-correction?
- pred_corr_lower_bnd
lower bound for the prediction-correction
- pi
simulated prediction interval to plot. Default is c(0.05, 0.95),
- ci
confidence interval to plot. Default is (0.05, 0.95)
- uloq
Number or NULL indicating upper limit of quantification. Default is NULL.
- lloq
Number or NULL indicating lower limit of quantification. Default is NULL.
- log_y
Boolean indicting whether y-axis should be shown as logarithmic. Default is FALSE.
- log_y_min
minimal value when using log_y argument. Default is 1e-3.
- xlab
label for x axis
- ylab
label for y axis
- title
title
- smooth
"smooth" the VPC (connect bin midpoints) or show bins as rectangular boxes. Default is TRUE.
- vpc_theme
theme to be used in VPC. Expects list of class vpc_theme created with function vpc_theme()
- facet
either "wrap", "columns", or "rows"
- scales
Are scales shared across all facets (the default,
"fixed"), or do they vary across rows ("free_x"), columns ("free_y"), or both rows and columns ("free")?- labeller
ggplot2 labeller function to be passed to underlying ggplot object
- vpcdb
Boolean whether to return the underlying vpcdb rather than the plot
- verbose
show debugging information (TRUE or FALSE)
- ...
Args sent to
rxSolve- seed
an object specifying if and how the random number generator should be initialized
- idv
Name of independent variable. For
vpcPlot()andvpcCens()the default is"time"forvpcPlotTad()andvpcCensTad()this is"tad"- cens
is a boolean to show if this is a censoring plot or not. When
cens=TRUEthis is actually a censoring vpc plot (withvpcCens()andvpcCensTad()). Whencens=FALSEthis is traditional VPC plot (vpcPlot()andvpcPlotTad()).- method
the method to use for VPC plotting; can be
"vpc"(uses the vpc package) or"tidyvpc"(uses the tidyvpc package). By default,"vpc"is used when the vpc package is available, otherwise"tidyvpc"is used.
Examples
# \donttest{
one.cmt <- function() {
ini({
tka <- 0.45; label("Ka")
tcl <- log(c(0, 2.7, 100)); label("Cl")
tv <- 3.45; label("V")
eta.ka ~ 0.6
eta.cl ~ 0.3
eta.v ~ 0.1
add.sd <- 0.7; label("Additive residual error")
})
model({
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
linCmt() ~ add(add.sd)
})
}
fit <-
nlmixr2est::nlmixr(
one.cmt,
data = nlmixr2data::theo_sd,
est = "saem",
control = list(print = 0)
)
#>
#>
#>
#>
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#>
#> → loading into symengine environment...
#> → pruning branches (`if`/`else`) of saem model...
#> ✔ done
#> → finding duplicate expressions in saem model...
#> ✔ done
#> ℹ calculate uninformed etas
#> ℹ done
#> Calculating covariance matrix
#> → loading into symengine environment...
#> → pruning branches (`if`/`else`) of saem model...
#> ✔ done
#> → finding duplicate expressions in saem predOnly model 0...
#> → finding duplicate expressions in saem predOnly model 1...
#> → finding duplicate expressions in saem predOnly model 2...
#> ✔ done
#>
#>
#> → Calculating residuals/tables
#> ✔ done
#> → compress origData in nlmixr2 object, save 6584
#> → compress parHistData in nlmixr2 object, save 8280
#> → compress phiM in nlmixr2 object, save 312896
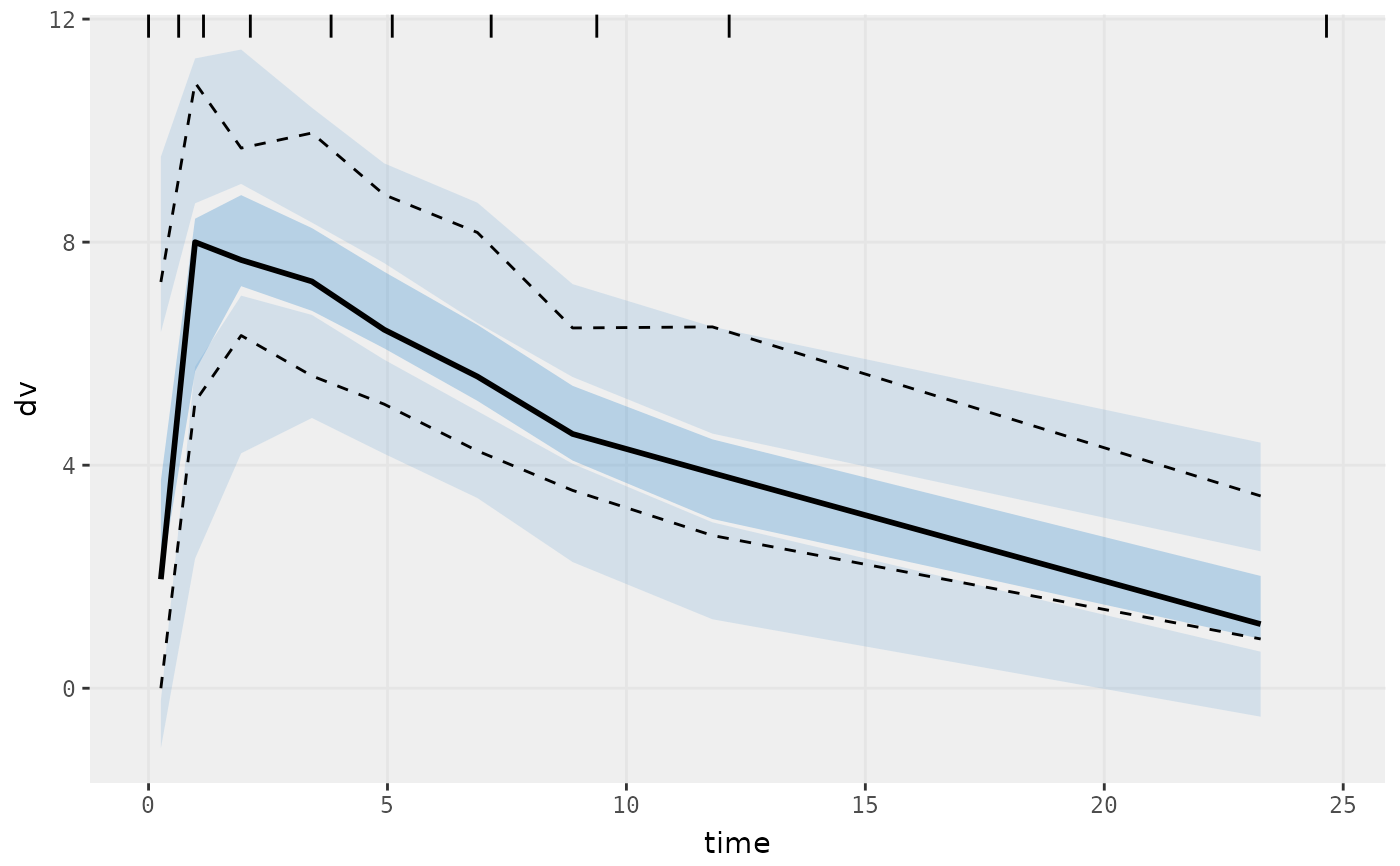
vpcPlot(fit)
#>
#>
#> Warning: No software packages matched for filtering values, not filtering.
#> Object class: other, data.frame
#> Available filters: phoenix, nonmem
#> Warning: No software packages matched for filtering values, not filtering.
#> Object class: other, nlmixr2vpcSim, data.frame
#> Available filters: phoenix, nonmem
 # }
# }
