Plot some standard goodness of fit plots for the focei fitted object
Usage
# S3 method for class 'nlmixr2FitData'
plot(x, ...)Value
An nlmixr2PlotList object (a list of ggplot2 objects with easier
plotting for all of them at the same time)
Examples
# \donttest{
library(nlmixr2est)
one.compartment <- function() {
ini({
tka <- 0.45
tcl <- 1
tv <- 3.45
eta.ka ~ 0.6
eta.cl ~ 0.3
eta.v ~ 0.1
add.sd <- 0.7
})
model({
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
d/dt(depot) = -ka * depot
d/dt(center) = ka * depot - cl / v * center
cp = center / v
cp ~ add(add.sd)
})
}
## The fit is performed by the function nlmixr/nlmix2 specifying the model, data and estimate
fit <- nlmixr2(one.compartment, theo_sd, est="saem", saemControl(print=0, nBurn = 10, nEm = 20))
#>
#>
#>
#>
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#>
#> → loading into symengine environment...
#> → pruning branches (`if`/`else`) of saem model...
#> ✔ done
#> → finding duplicate expressions in saem model...
#> → optimizing duplicate expressions in saem model...
#> ✔ done
#> ℹ calculate uninformed etas
#> ℹ done
#> Calculating covariance matrix
#> → loading into symengine environment...
#> → pruning branches (`if`/`else`) of saem model...
#> ✔ done
#> → finding duplicate expressions in saem predOnly model 0...
#> → finding duplicate expressions in saem predOnly model 1...
#> → optimizing duplicate expressions in saem predOnly model 1...
#> → finding duplicate expressions in saem predOnly model 2...
#> ✔ done
#>
#>
#> → Calculating residuals/tables
#> ✔ done
#> → compress origData in nlmixr2 object, save 6584
#> → compress parHistData in nlmixr2 object, save 2560
#> → compress phiM in nlmixr2 object, save 15080
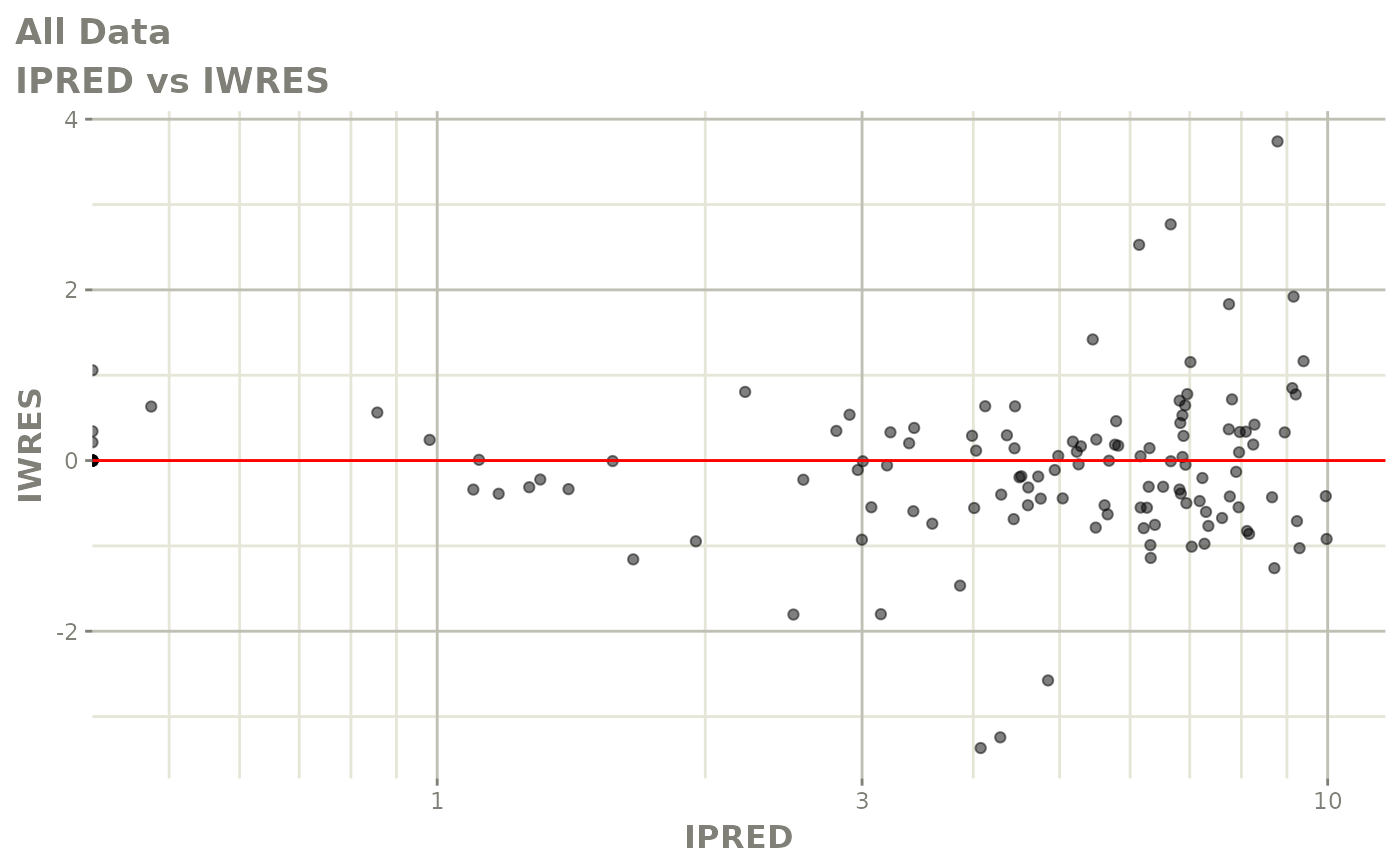
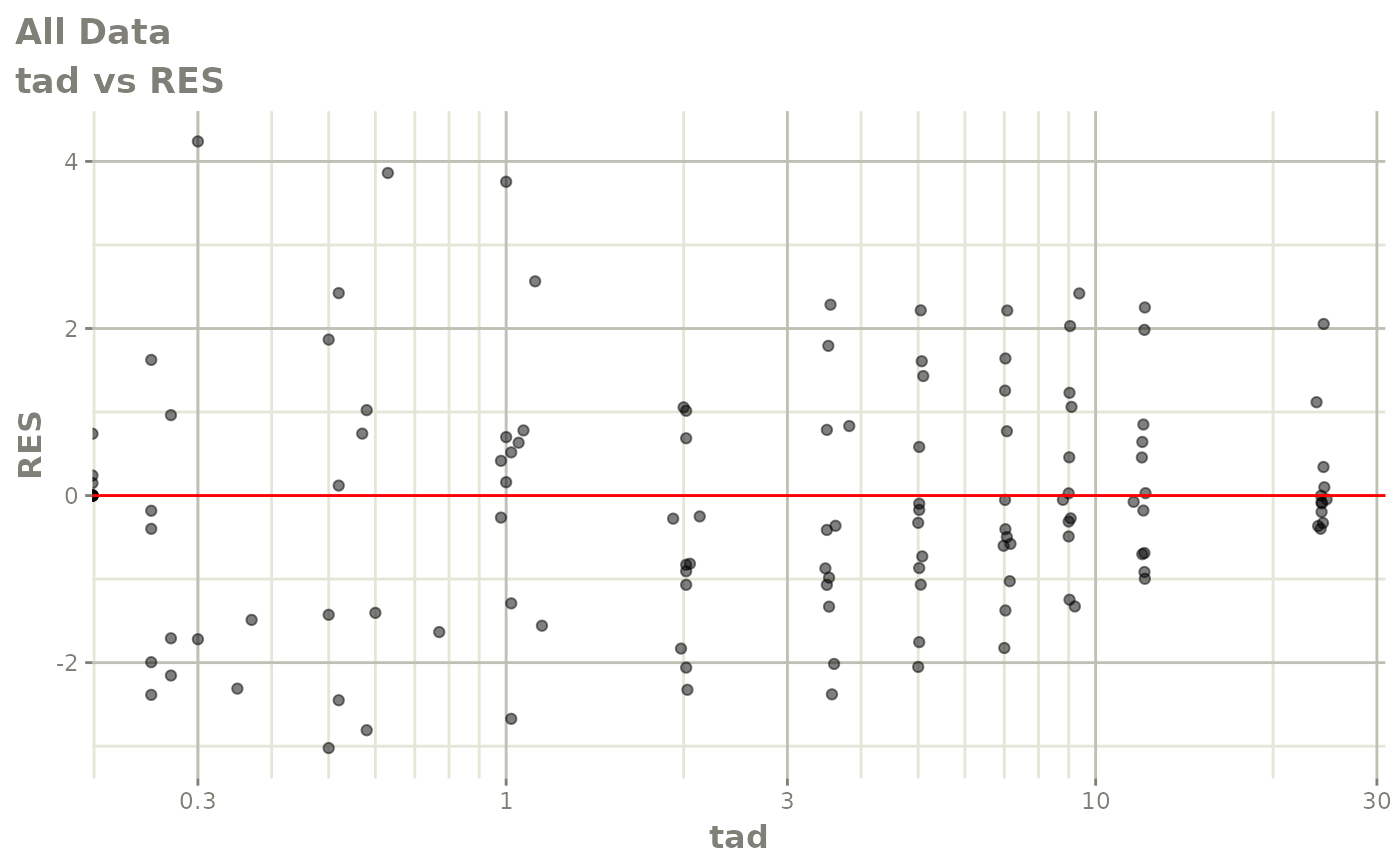
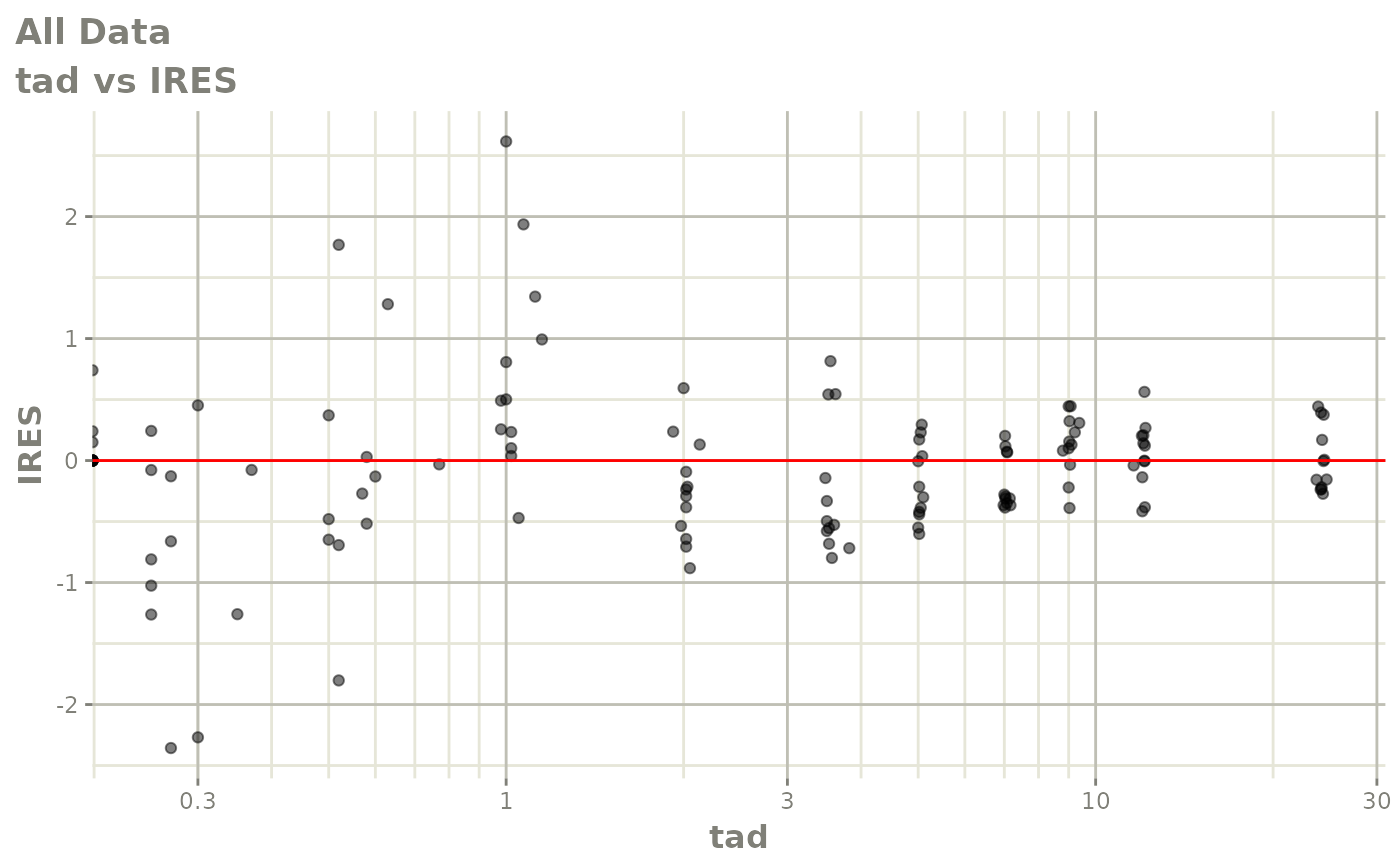
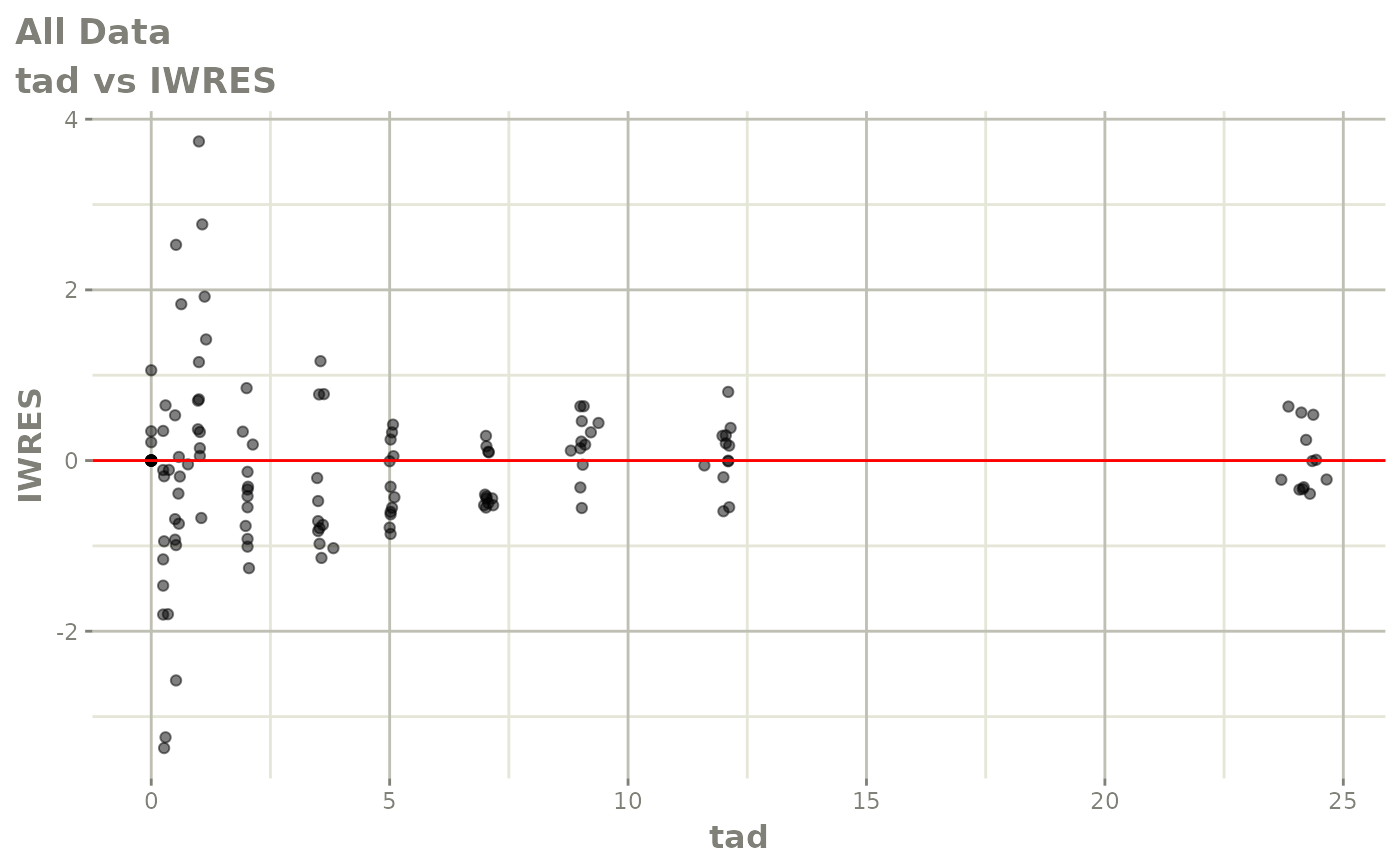
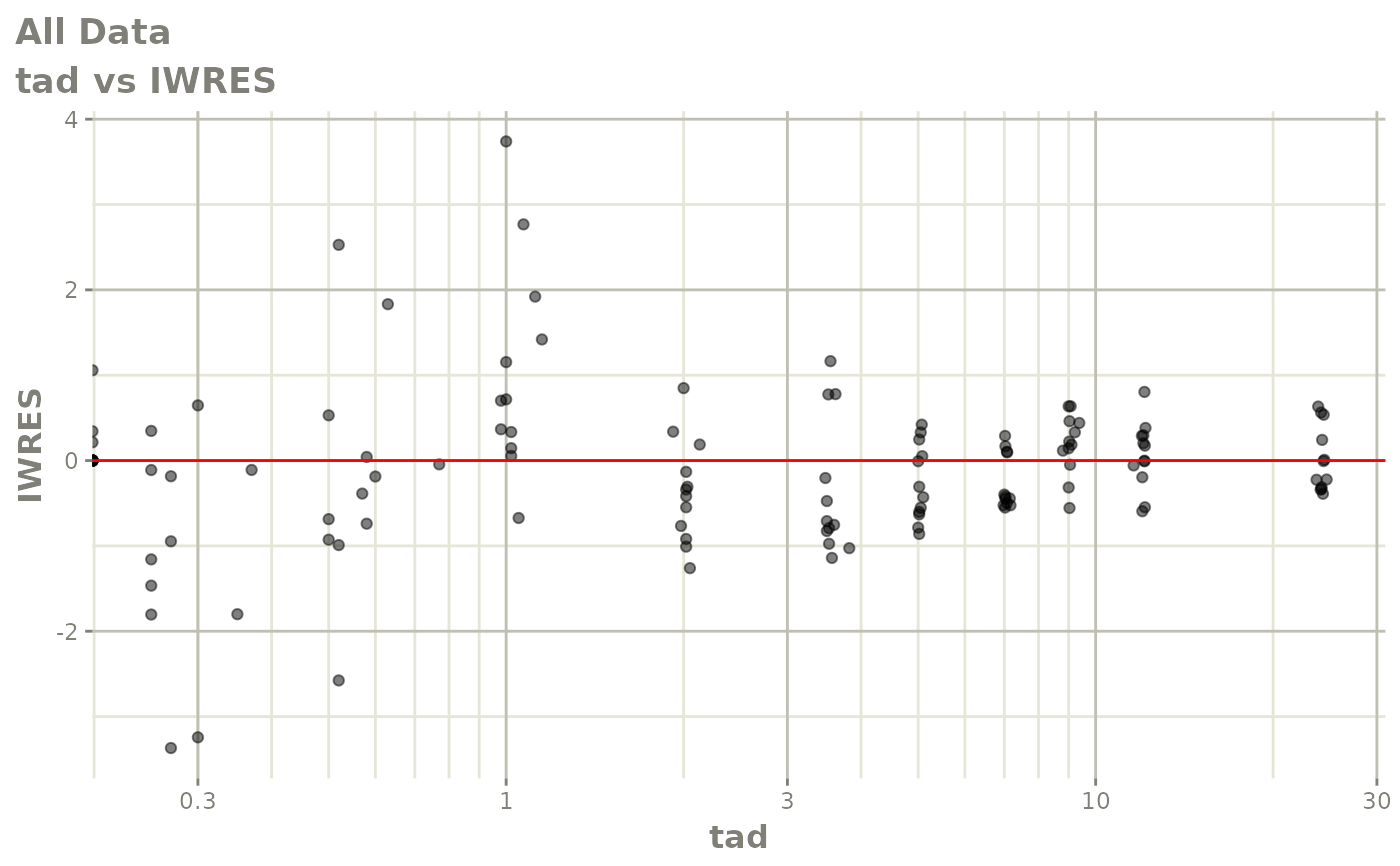
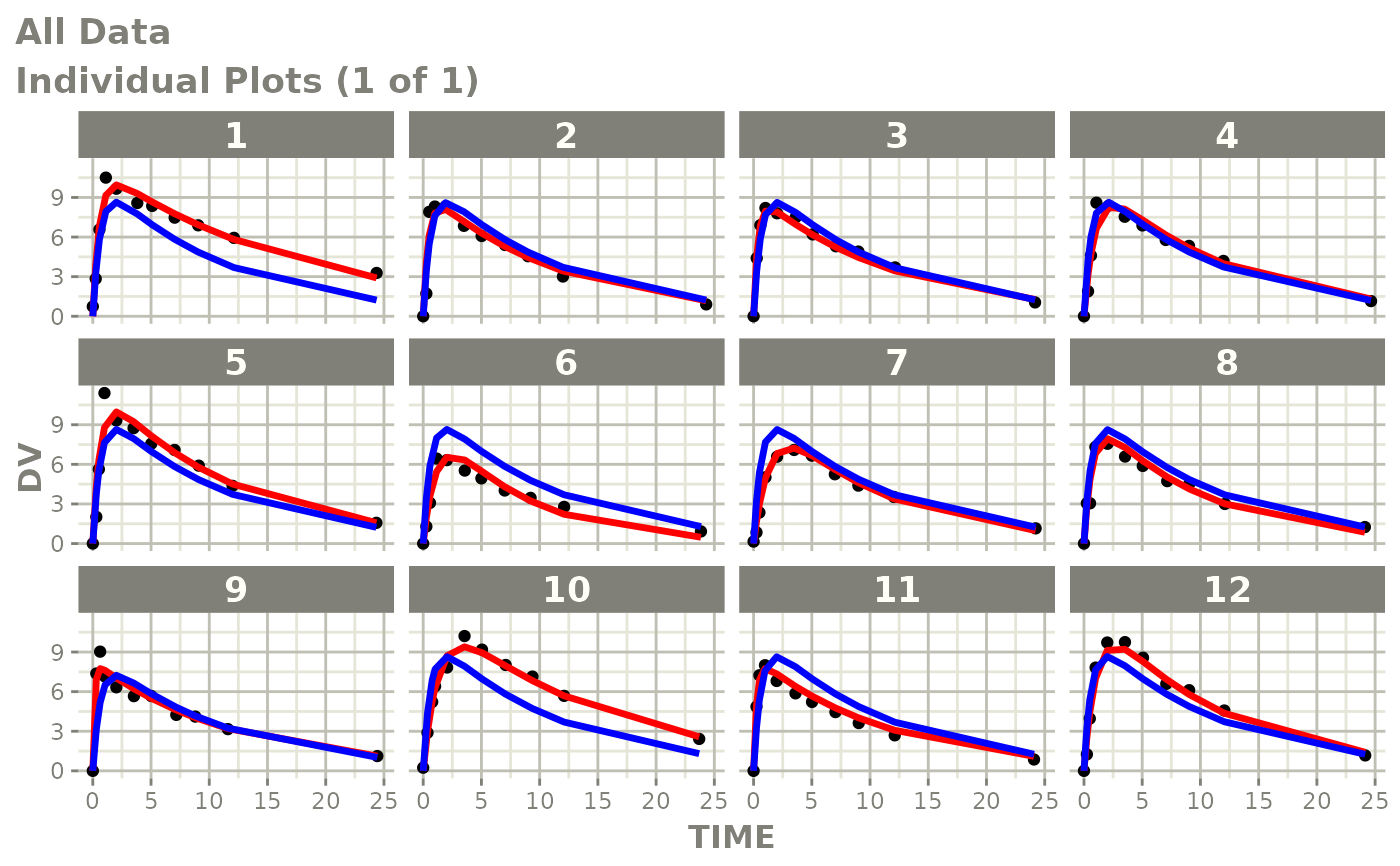
# This shows many goodness of fit plots
plot(fit)
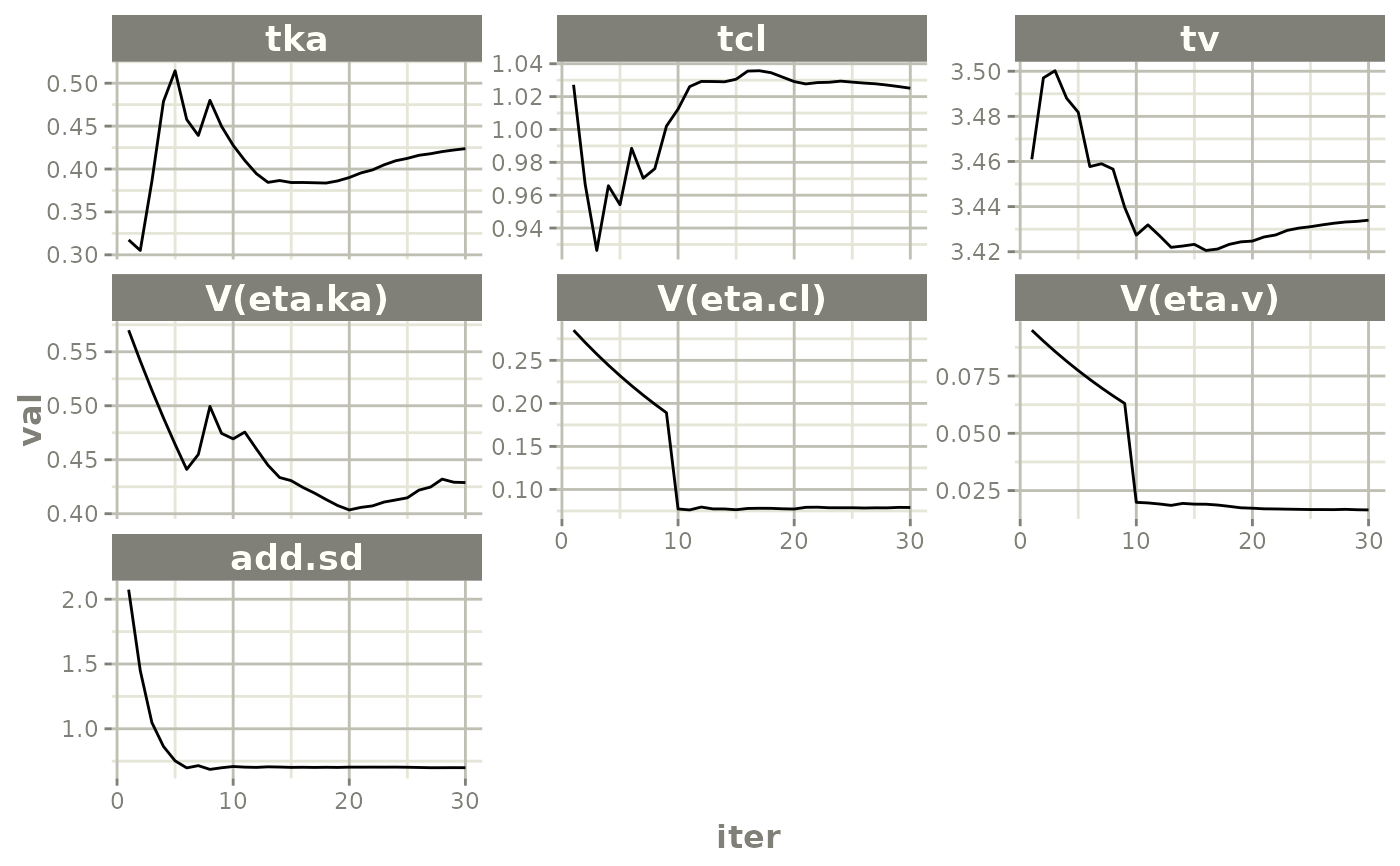
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
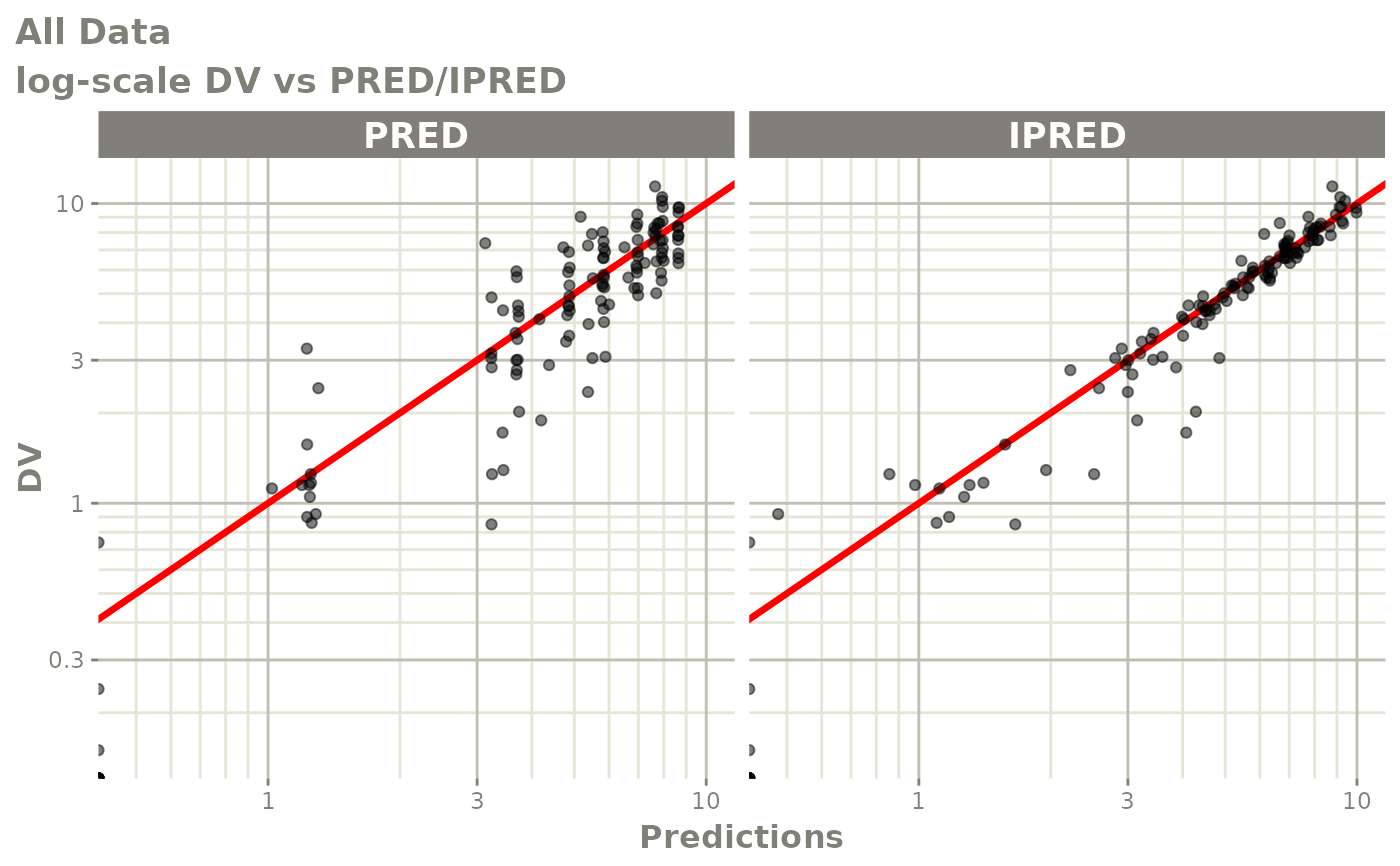
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
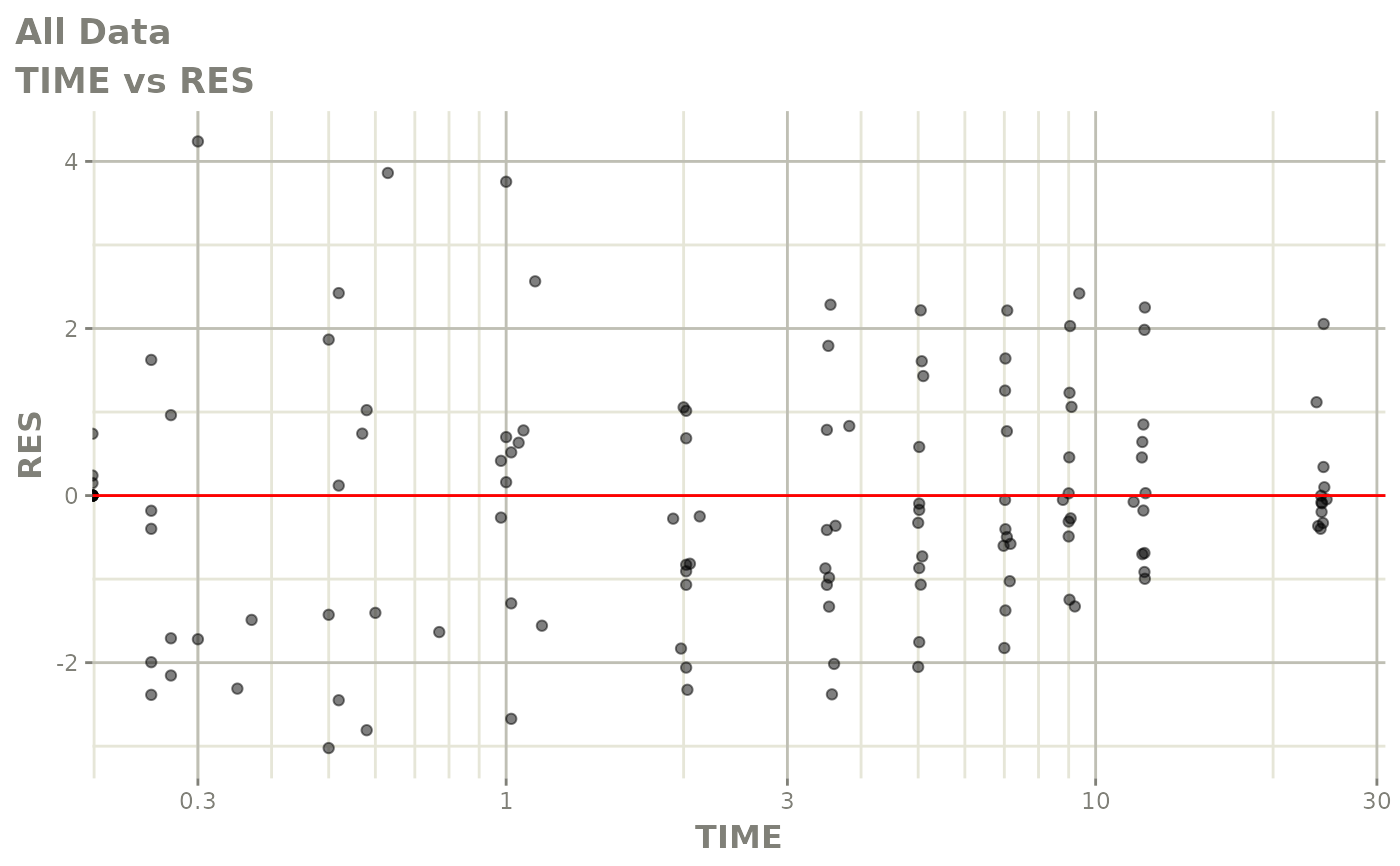
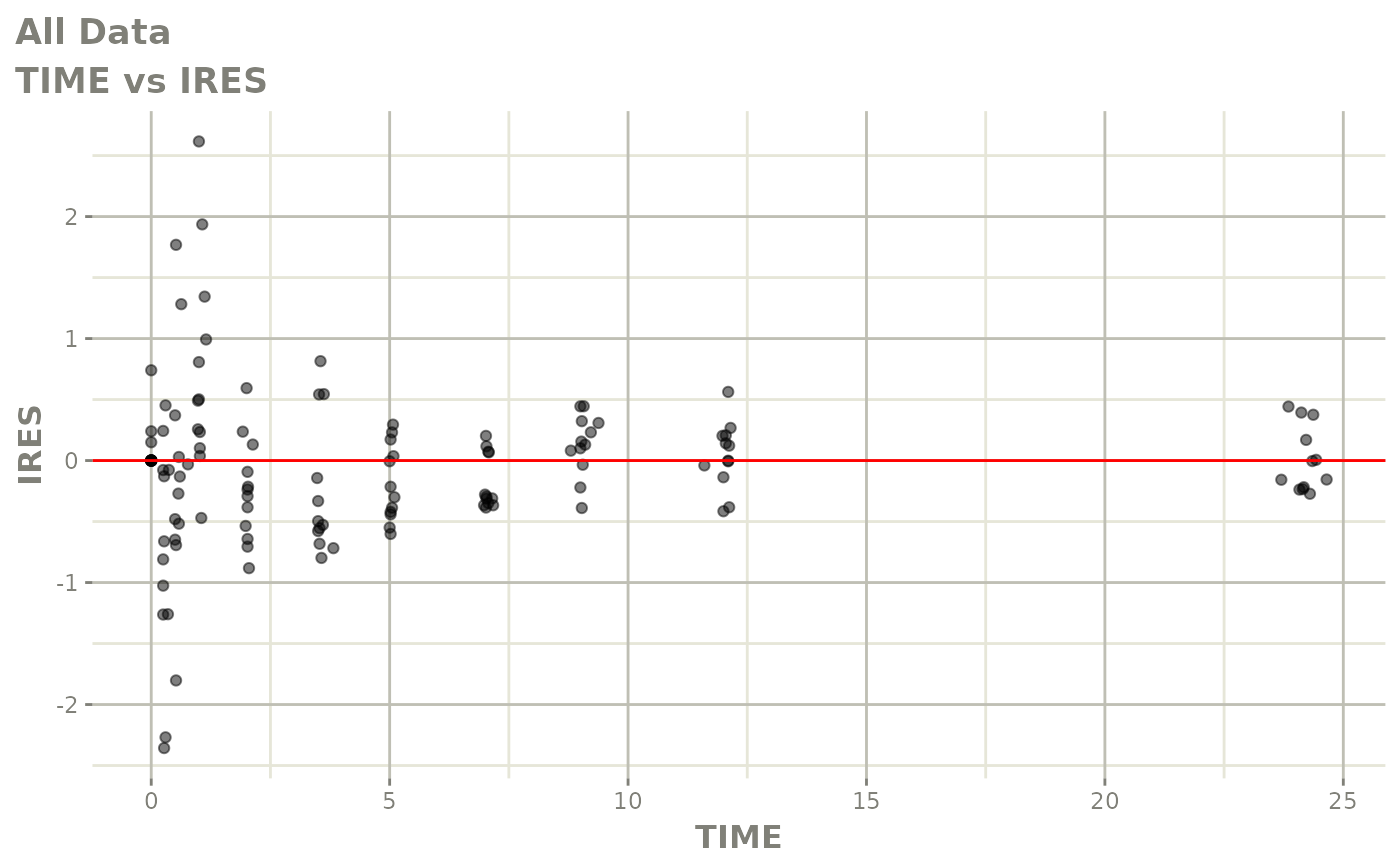 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
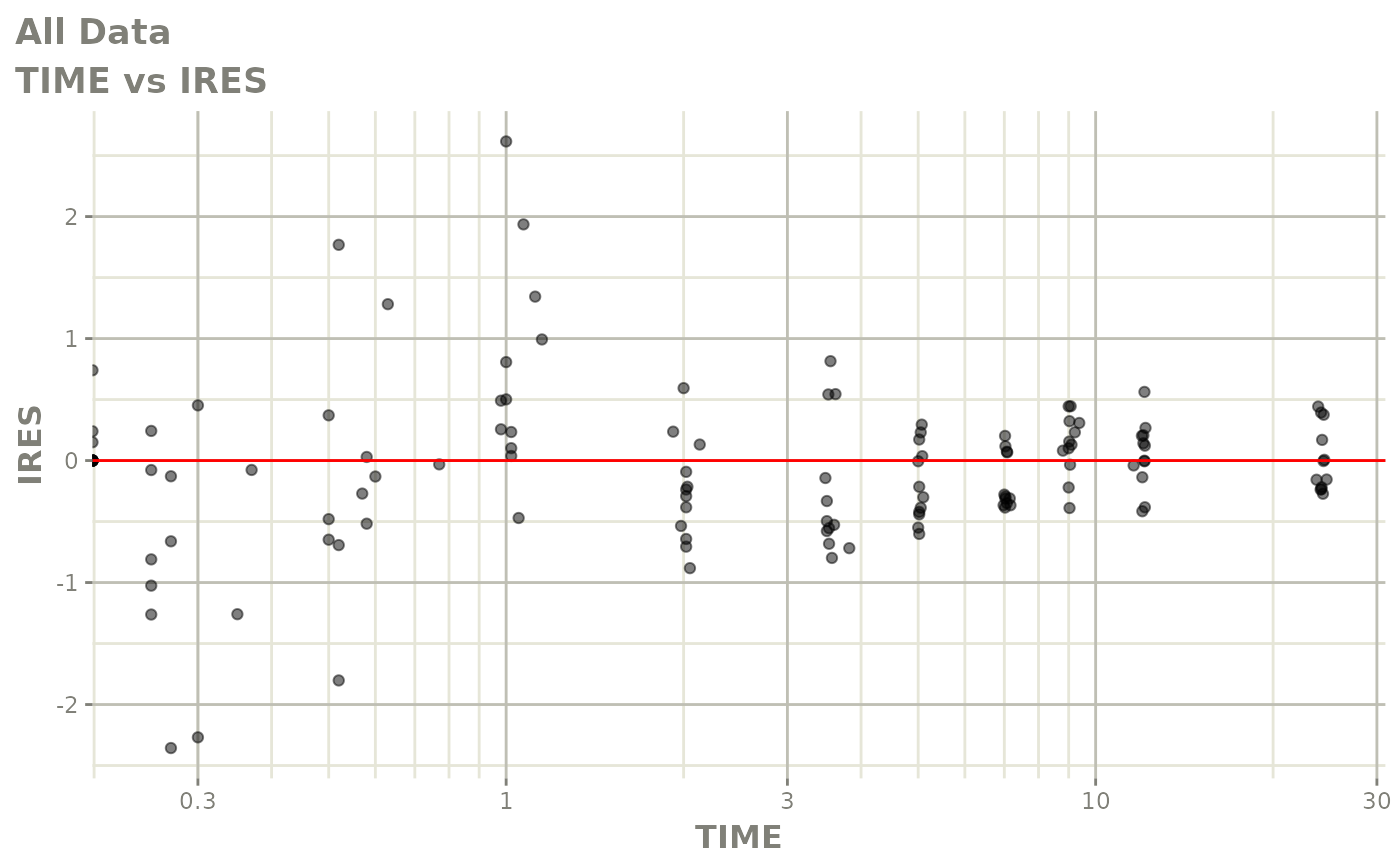
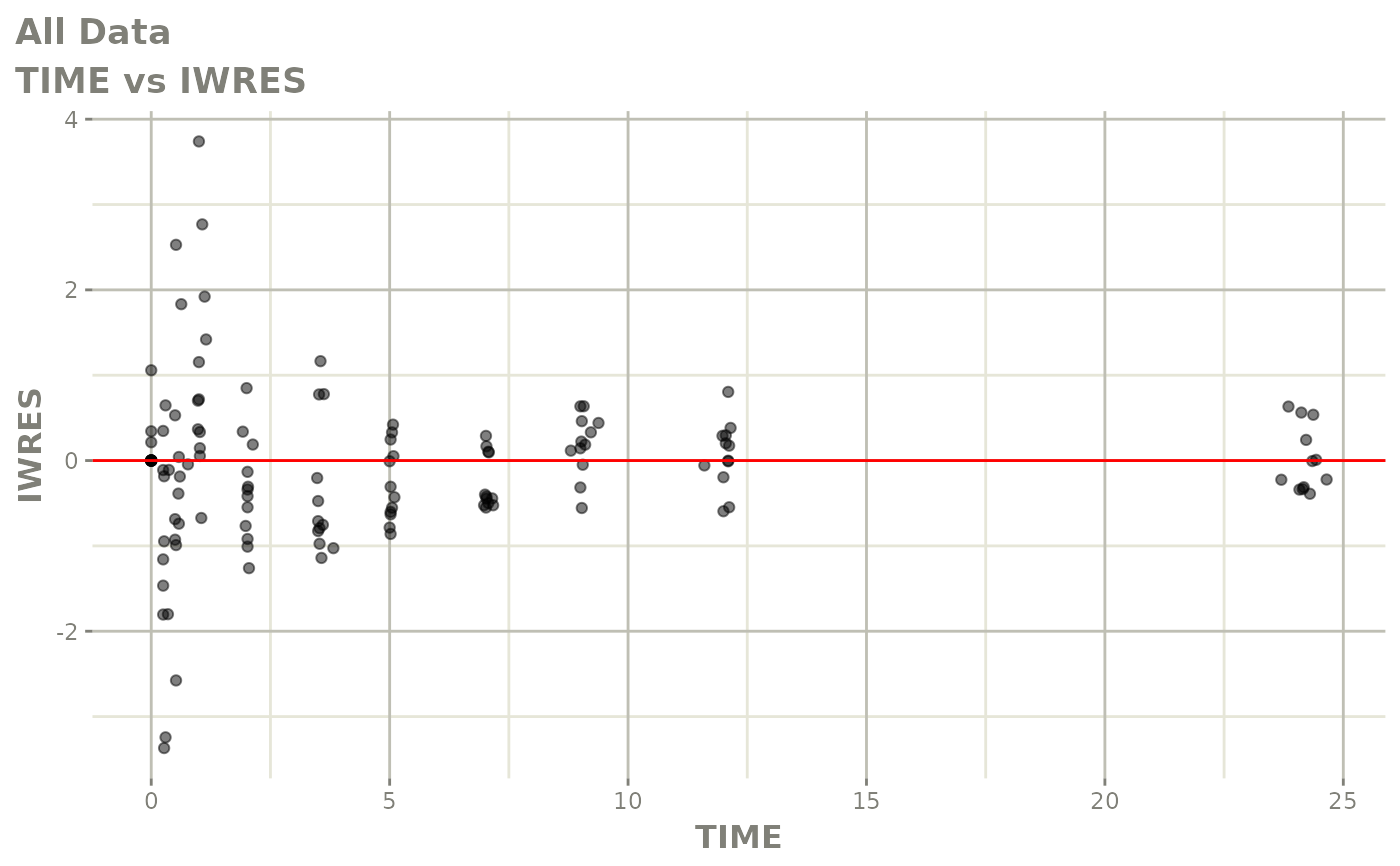 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.

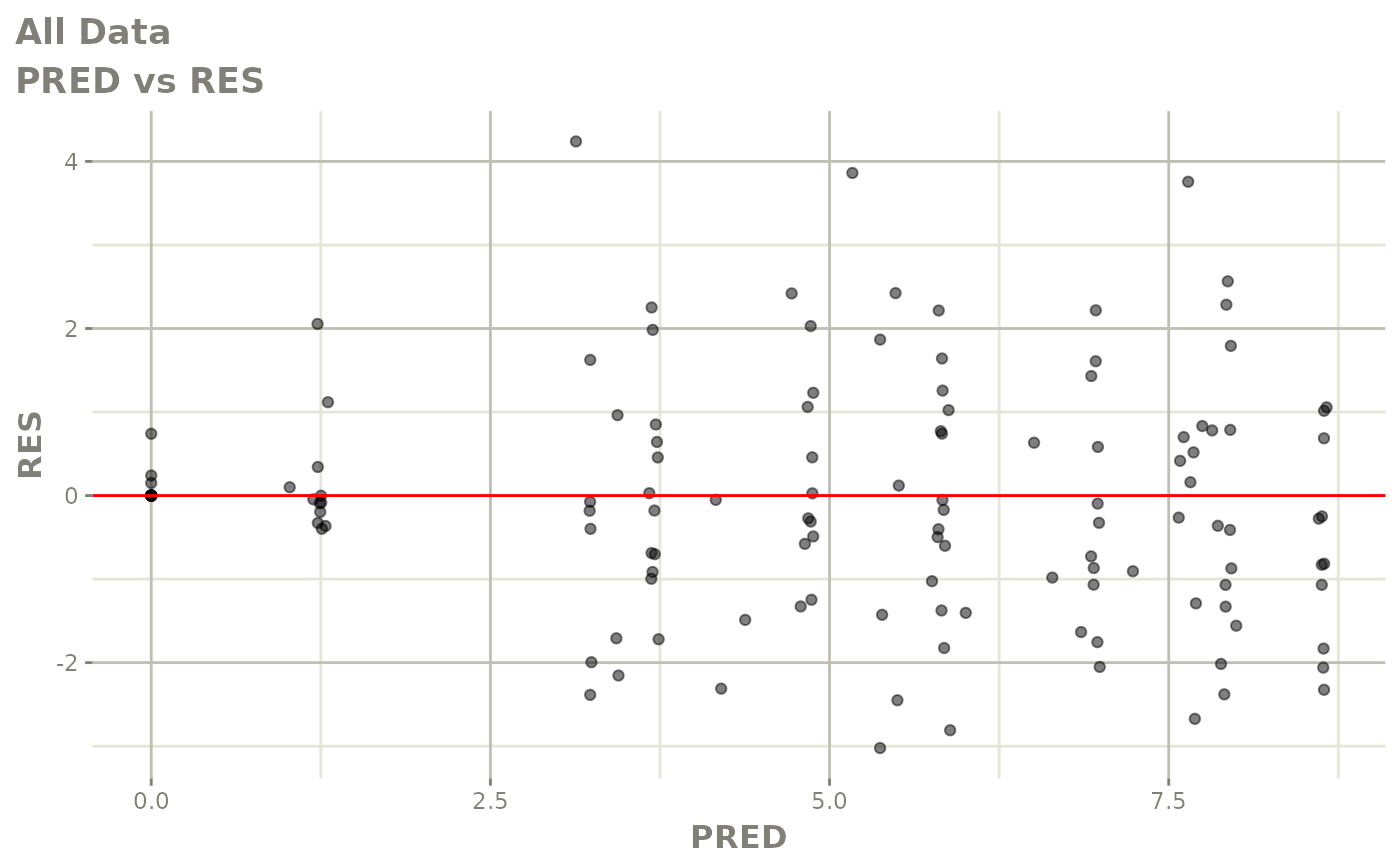 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
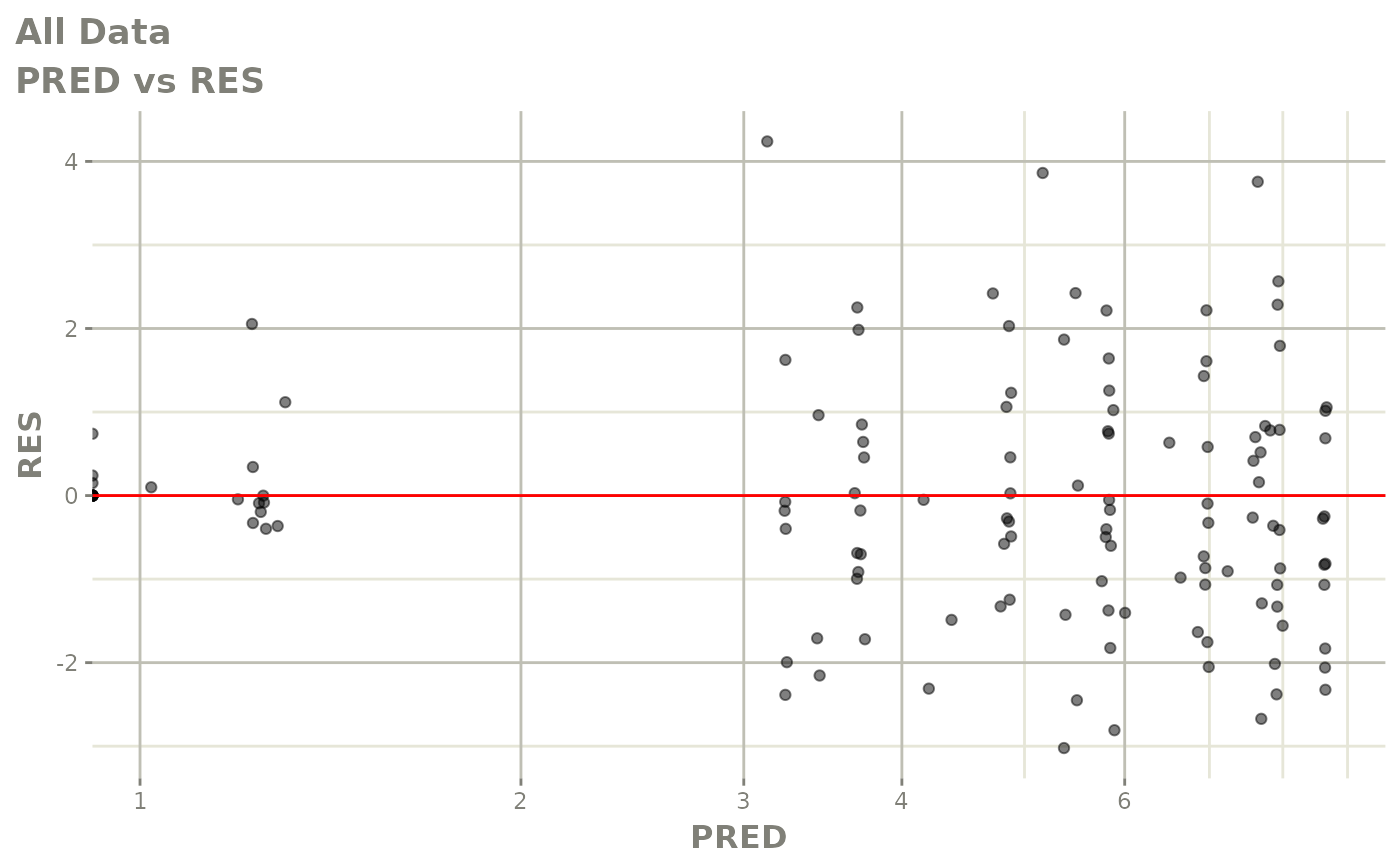
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
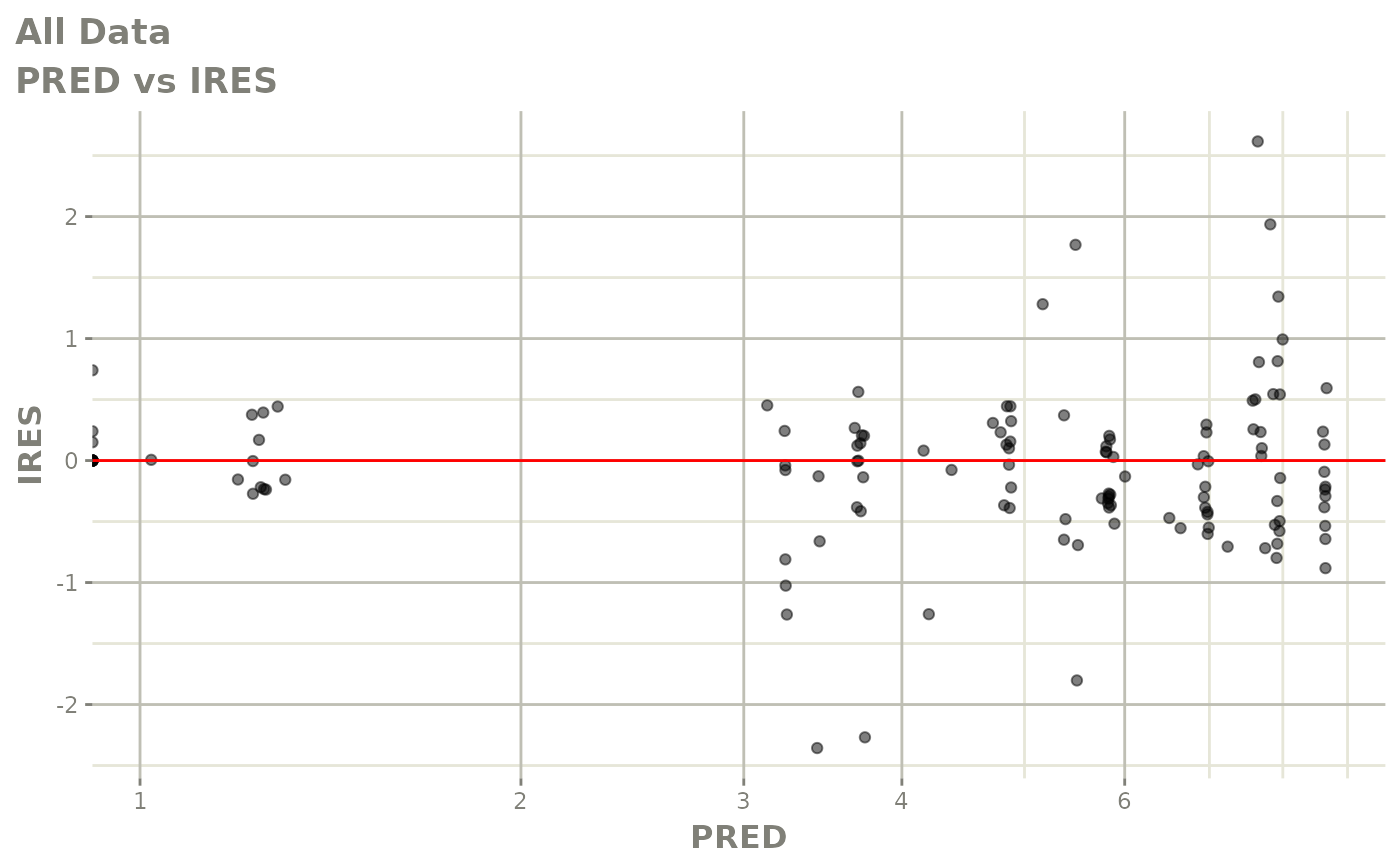
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
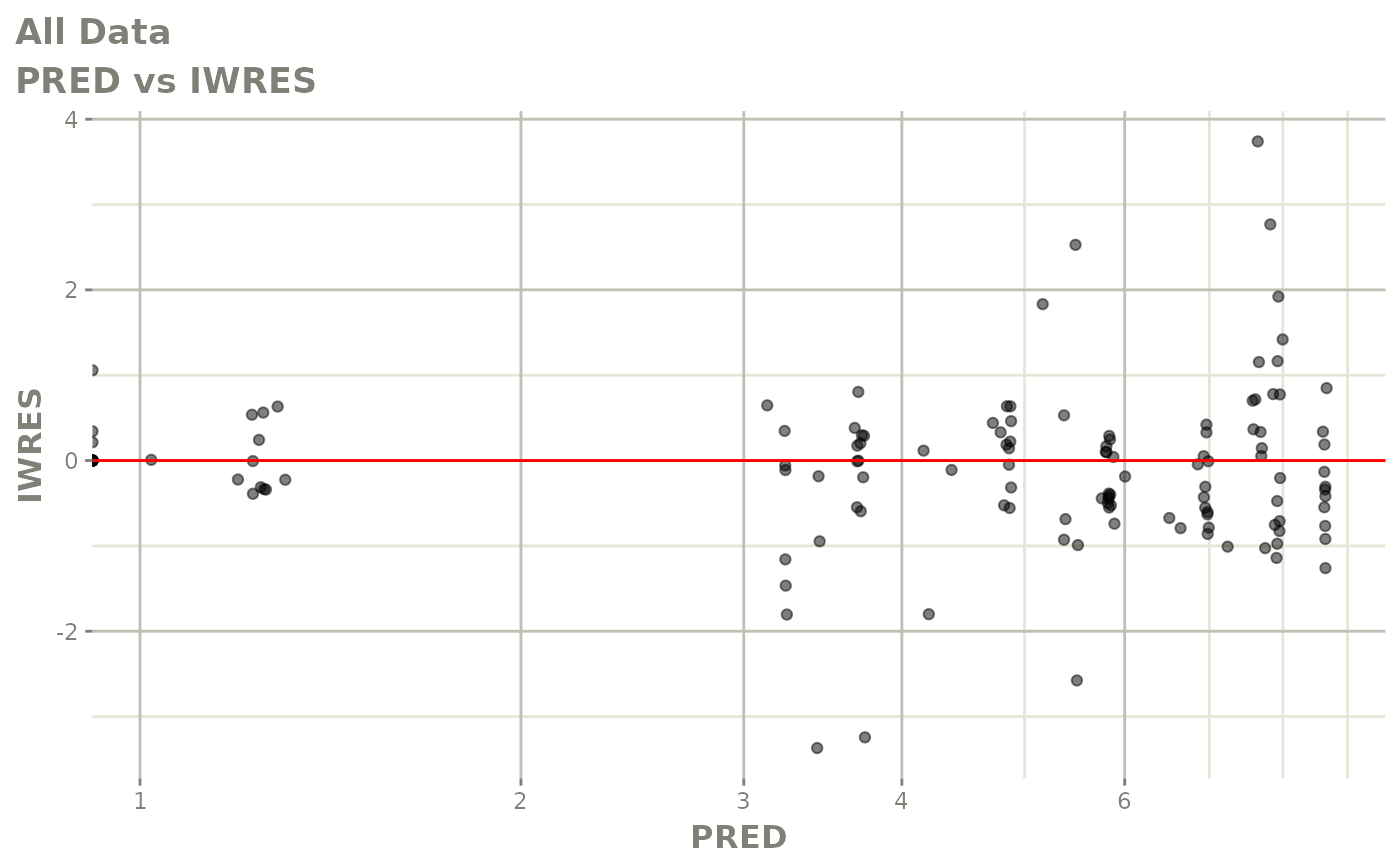
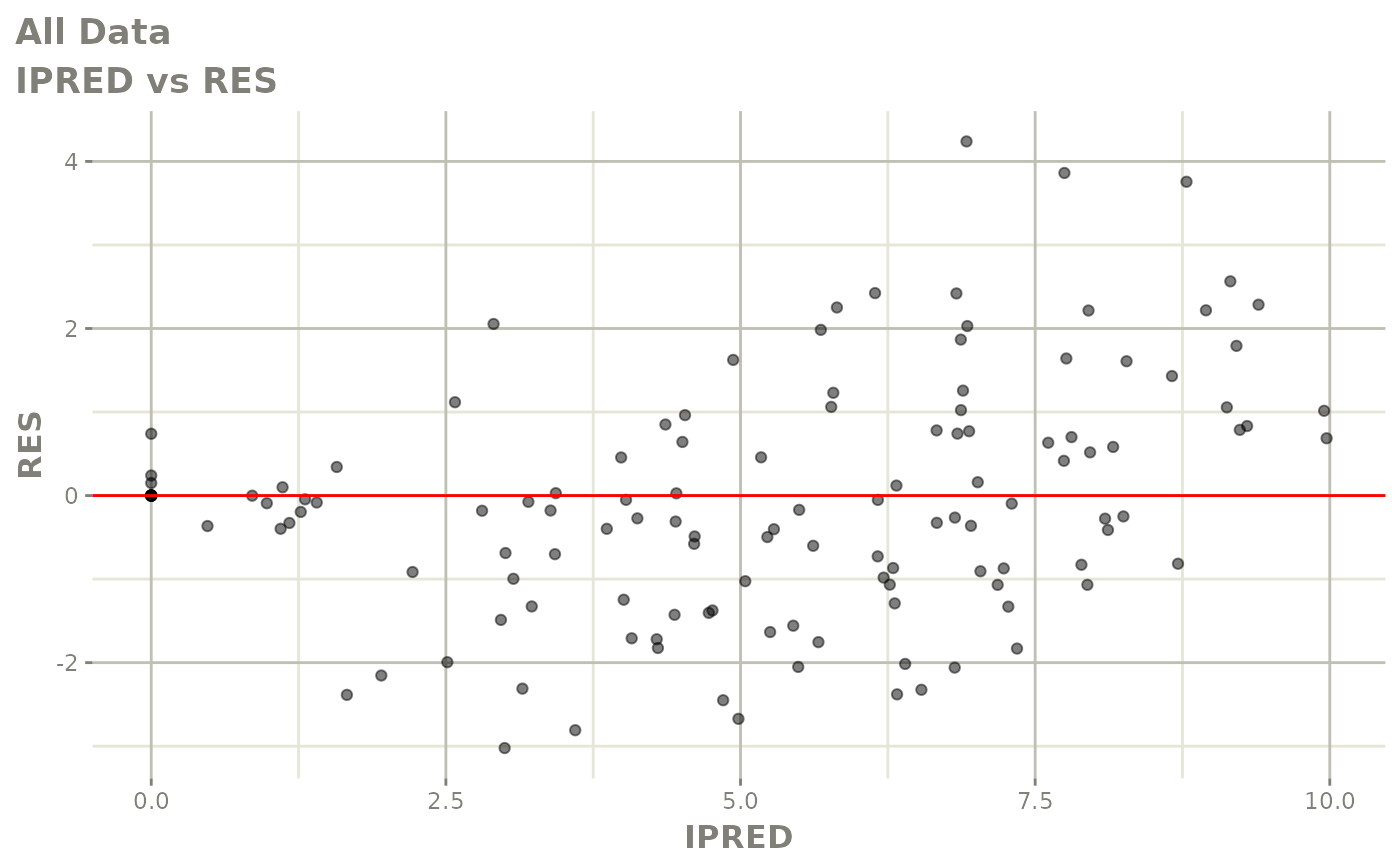 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
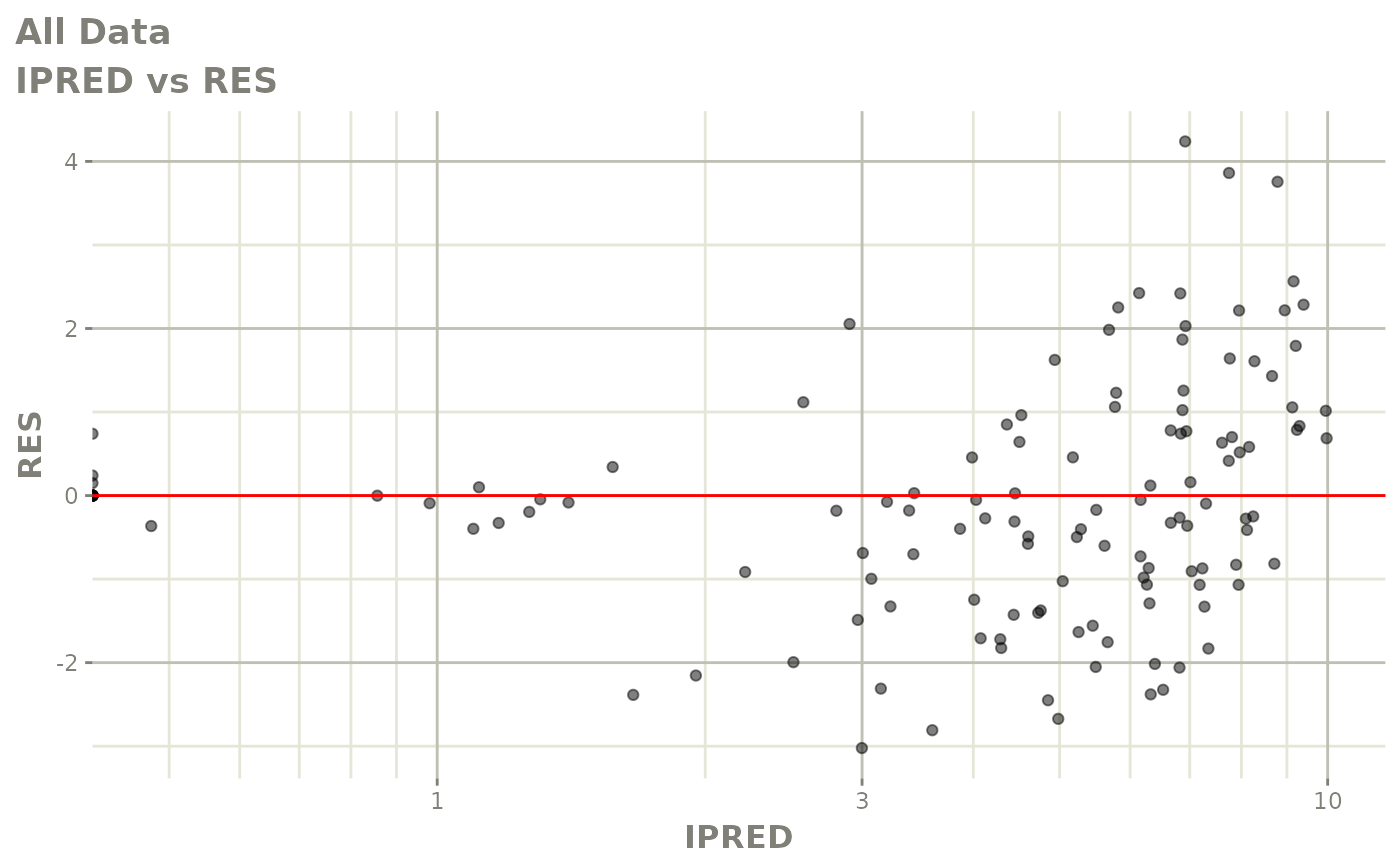
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.

 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
 #> Warning: log-10 transformation introduced infinite values.
#> Warning: log-10 transformation introduced infinite values.
 # }
# }
