Plot a nlmixr2 augPred object
Usage
# S3 method for class 'nlmixr2AugPred'
plot(x, y, ...)Examples
# \donttest{
library(nlmixr2est)
#> Loading required package: nlmixr2data
## The basic model consiss of an ini block that has initial estimates
one.compartment <- function() {
ini({
tka <- 0.45 # Log Ka
tcl <- 1 # Log Cl
tv <- 3.45 # Log V
eta.ka ~ 0.6
eta.cl ~ 0.3
eta.v ~ 0.1
add.sd <- 0.7
})
# and a model block with the error sppecification and model specification
model({
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
d/dt(depot) = -ka * depot
d/dt(center) = ka * depot - cl / v * center
cp = center / v
cp ~ add(add.sd)
})
}
## The fit is performed by the function nlmixr/nlmix2 specifying the model, data and estimate
fit <- nlmixr2est::nlmixr2(one.compartment, theo_sd, est="saem", saemControl(print=0))
#>
#>
#>
#>
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#>
#> → loading into symengine environment...
#> → pruning branches (`if`/`else`) of saem model...
#> ✔ done
#> → finding duplicate expressions in saem model...
#> → optimizing duplicate expressions in saem model...
#> ✔ done
#> ℹ calculate uninformed etas
#> ℹ done
#> rxode2 5.0.1.9000 using 2 threads (see ?getRxThreads)
#> no cache: create with `rxCreateCache()`
#>
#> Attaching package: ‘rxode2’
#> The following objects are masked from ‘package:nlmixr2est’:
#>
#> boxCox, yeoJohnson
#> Calculating covariance matrix
#> → loading into symengine environment...
#> → pruning branches (`if`/`else`) of saem model...
#> ✔ done
#> → finding duplicate expressions in saem predOnly model 0...
#> → finding duplicate expressions in saem predOnly model 1...
#> → optimizing duplicate expressions in saem predOnly model 1...
#> → finding duplicate expressions in saem predOnly model 2...
#> ✔ done
#>
#>
#> → Calculating residuals/tables
#> ✔ done
#> → compress origData in nlmixr2 object, save 6584
#> → compress parHistData in nlmixr2 object, save 8344
#> → compress phiM in nlmixr2 object, save 312064
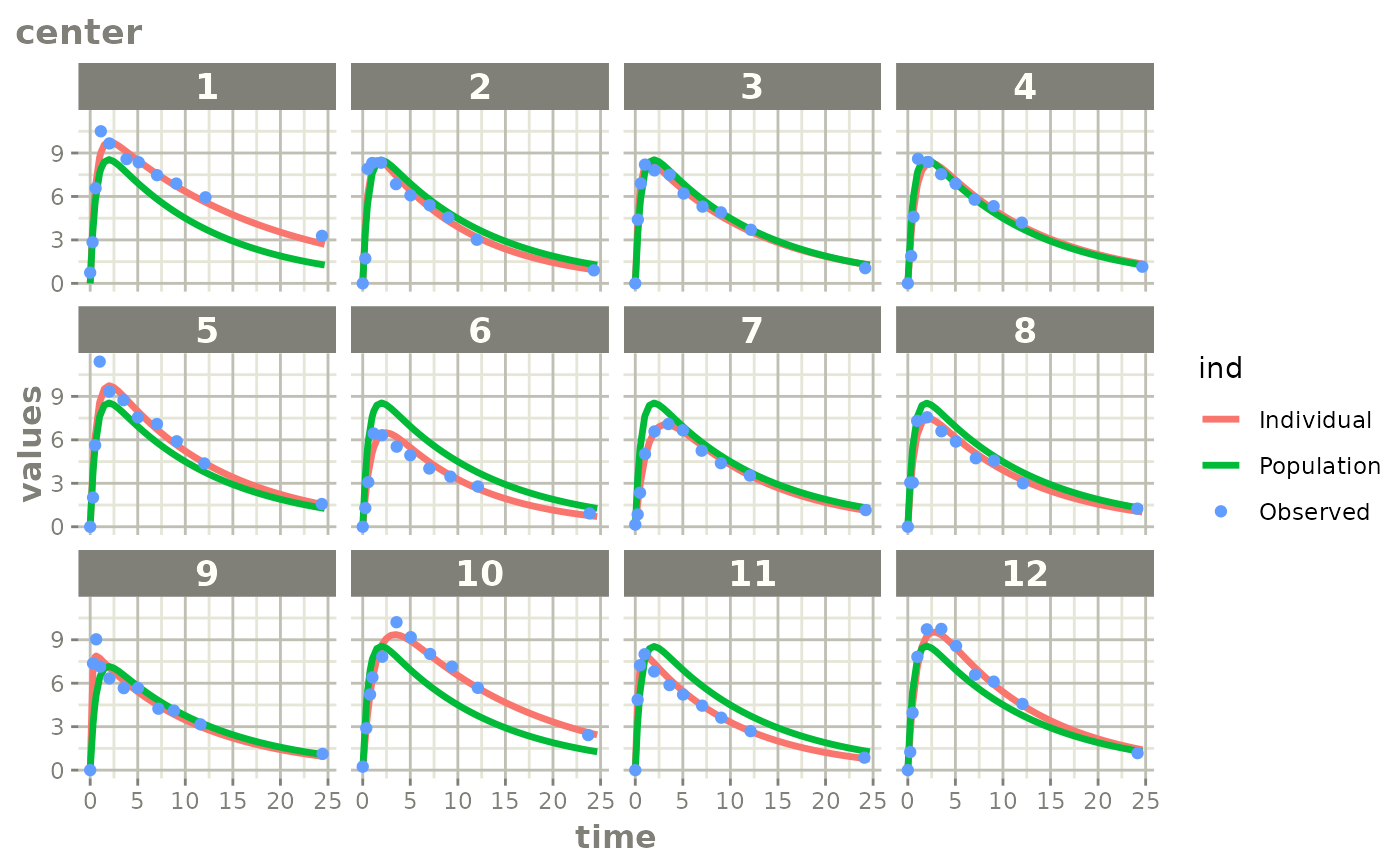
# augPred shows more points for the fit:
a <- nlmixr2est::augPred(fit)
#>
#>
# you can plot it with plot(augPred object)
plot(a)
 # }
# }
